Associations between soil microbial communities, gut microbiome, and Child Health in Kenya
My doctoral dissertation research addresses the relationship between the local soil microbiome and human gut microbiota for children in rural households in Kenya.
Prior research into this area supports the notion that the microbes around us affect the microbes within us, and this work contributes new evidence from a relatively large, novel cohort of children and households to help elucidate whether and to what extent microbes in soils–the most diverse microbial reservoir that exists–contribute to patterns in gut microbiome composition and structure.
The human gut microbiome has been identified as a strong modulator of immune and metabolic function, and consequently influences many dimensions of health and disease. Therefore, greater understanding of how the gut microbiome is influenced by, and responds to, its environment has the potential for unlocking new knowledge about the environmental drivers of health. This work is particularly salient for rural communities in East Africa, where burdens of communicable disease remain persistently high, and non-communicable diseases are on the rise.

Household outdoor environment in a rural home, western Kenya. Photo: Mariah Coley
Funding
◊ NIH Fogarty / UC Global Health Institute GloCal Health Fellowship
◊ Henry A. Jastro Research Award

Sylvia Akoko, research assistant, training in our surface soil sample collection protocol. Photo: Mariah Coley
Field sample and data collection
I led a small team in carrying out data and sample collection in 144 rural households in western Kenya over 6 months in 2022. Data collected include:
- A fecal sample from the child participant
- 3 surface soil samples from the child’s home environment
- A household questionnaire with the child’s primary caregiver
- A scorecard measuring the presence and severity of recent diarrheal episodes for the child
- A sample the household’s point-of-consumption drinking water source
Sample processing
Fecal and soil samples were placed in cold storage at the laboratory of the Nyanza Reproductive Health Society at Lumumba Sub-County Hospital in Kisumu. We then used Qiagen microbial DNA extraction kits to isolate bacterial DNA from the fecal and soil samples.
After quantification, DNA was submitted to the Environmental Sample Preparation and Sequencing Facility at Argonne National Laboratory for 16S rRNA amplicon sequencing with Illumina MiSeq.
Drinking water samples were processed within 8 hours of collection using IDEXX Colilert and the Quanti-Tray system. After incubation, each tray was read for Most Probable Number (MPN) for total coliforms and E. Coli.
All participant and household questionnaire data was collated and stored in a secure cloud server prior to downstream analysis.

DNA extraction carried out at the Nyanza Reproductive Health Services laboratory in Kisumu. Photo: Mariah Coley
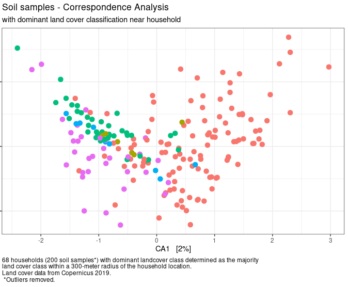
Exploratory ordination analysis with soils data.
Data Analysis
16S data was returned from Argonne National Laboratory in early 2023, and analysis is ongoing. Data processing and bioinformatics approaches used to date include:
- Barcode demultiplexing with idemp
- Data preprocessing with DADA2 pipeline (HPC cluster)
- Taxonomy assignment with DECIPHER and SILVA (HPC cluster)
- Ordination analyses of fecal and soil microbial communities with phyloseq and DESeq2
- Microbial source tracking via FEAST
- Integration of additional environmental, soils, and landscape data from Copernicus, CHIRPS, and iSDAsoil
All analyses are implemented in R.
Research team and collaborators
Principal Investigator: Mariah Coley
- Study conceptualization and design
- Funding acquisition
- IRB / ethical review preparation (UC Davis and Kenya Medical Research Institute)
- Field and laboratory protocol development, logistics and operations planning
- Full project budgeting and financial management
- Research team training
- Field data and sample collection
- Laboratory sample processing
- Bioinformatics and data analysis
- Manuscript prep
Advisors and mentors
- Kate Scow (PhD major advisor, UC Davis)
- Phelgona Otieno (mentor, Kenya Medical Research Institute)
- Craig Cohen (mentor, UC San Francisco)
- Robert Hijmans (dissertation committee, UC Davis)
Research Assistants
- Gloria Ger (Field coordination, field sample collection, laboratory support)
- Sylvia Akoko (Household questionnaire implementation)
- Perez Kitoto (Participant eligibility screening, field sample collection)
- Irene Oucho (Screening, questionnaire implementation, field sample collection)
Support
- Members of the Scow Lab, UC Davis
- Radomir Schmidt, Scow Lab at UC Davis
- Dr Fredrick Otieno, Nyanza Reproductive Health Society
- Walter Agingu, Nyanza Reproductive Health Society

From left: Sylvia, Perez, me, Gloria, and Irene.